Maikel Ronnau et al will publish Automatic segmentation and classification of Papanicolaou-stained cells and dataset for oral cancer detection in next month’s Computers in Biology and Medicine.
Their research uses a CNN U-Net architecture with ResNet encoder to achieve expert level performance by segmenting and classifying a trained on UFSC OCPap dataset of oral cytology images. The team uses transfer learning for image segmentation to train the model which is used to evaluate more than 1500 images from 52 patients labeled by specialists resulting in an average Dice score of 0.66 and Intersection over Union (IoU) of 0.65.
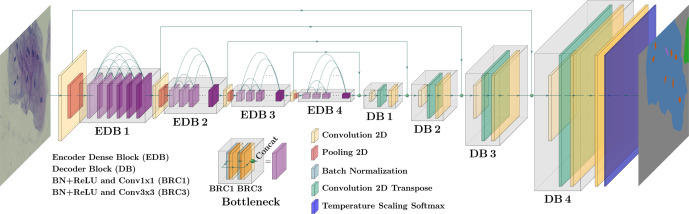
“The CNN model architecture for encoding layers are based on DenseNet-169 while the decoding layers are LinkNet but to replace the regular softmax layer with a temperature scaling softmax with a temperature parameter value of 0.1 to increase the confidence of the predictions and avoid the bias towards the prediction of background pixels. The model’s prediction is further processed by a semantic reclassification and a segmentation artifact removal steps.”
Findings suggest promising applications for computer functions in biology.



